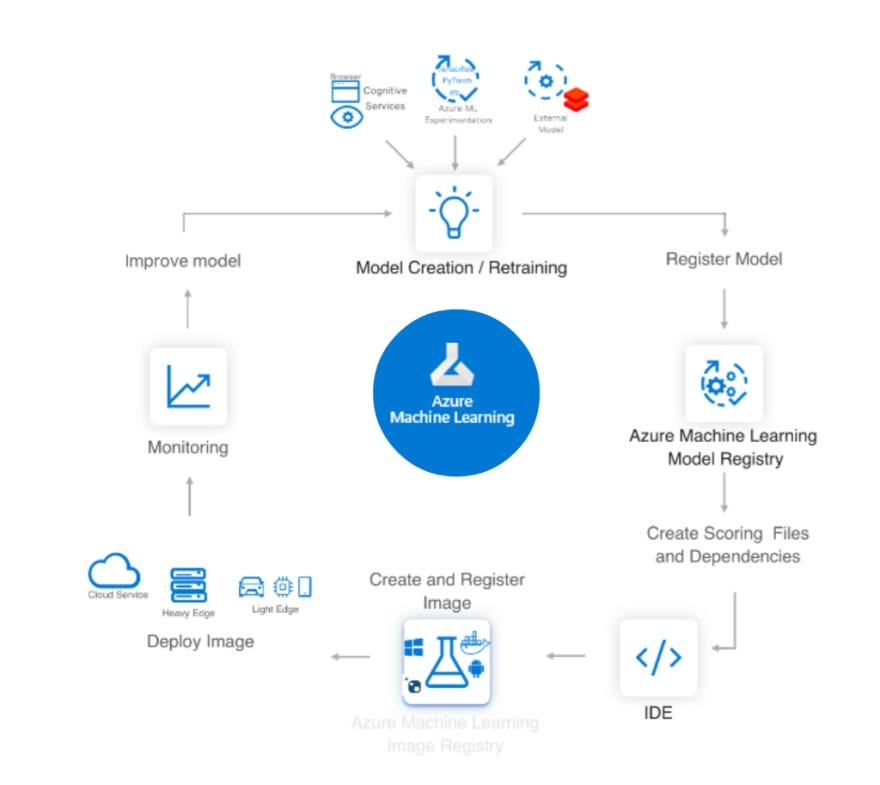
Since I did not find such numbers on both streaming services websites I decided to scrape full lists of movies and TV shows and calculate % of “fresh” / “old” movies in entire lists. Likely, there are different websites listing all movies and TV shows both on Disney+ and Netflix and updating them daily. I choose one based on popularity (I checked it both with Alexa and Similar Web) and its architecture making my scraping job easy and predictable.
library (tidyverse)
library (rvest)
disney0 <- read_html("https://www.finder.com/complete-list-disney-plus-movies-tv-shows-exclusives/")
Locate proper CSS element using Selector Gadget disney_year_0 <- html_nodes(disney0, 'td:nth-child(2)') disney_year <- html_text (disney_year_0, trim=TRUE) # list of release years for every title length (disney_year) #1029Now, let`s create frequency table for each year of release
disney_year_table <- as.data.frame (sort (table (disney_year), decreasing = TRUE), stringsAsFactors = FALSE)
colnames (disney_year_table) <- c('years', 'count')
glimpse (disney_year_table)
Rows: 89 Columns: 2 $ years "2019", "2017", "2016", "2018", "2015", "2011", "2014", "2012", "2010", "200... $ count 71, 58, 48, 40, 36, 35, 35, 34, 31, 28, 28, 27, 27, 25, 24, 24, 23, 23, 21,
Finally, count ‘fresh’ and 'old' % in entire Disney+ offer
disney_year_table$years <- disney_year_table$years %>% as.numeric () fresh_disney_years <- disney_year_table$years %in% c(2018:2020) fresh_disney_num <- paste (round (sum (disney_year_table$count[fresh_disney_years]) / sum (disney_year_table$count)*100),'%') old_disney_years <- disney_year_table$years < 2000 old_disney_num <- paste (round (sum (disney_year_table$count[old_disney_years]) / sum (disney_year_table$count)*100),'%')So, I`ve got roughly 14% “fresh” and 33% “old” movie on Disney+. Let`s see the numbers for Netflix.
For Netflix content our source website does not have single page so I scraped movies and TV shows separately.
#movies
netflix_mov0 <- read_html("https://www.finder.com/netflix-movies/")
netflix_mov_year_0 <- html_nodes(netflix_mov0, 'td:nth-child(2)')
netflix_mov_year <- tibble (html_text (netflix_mov_year_0, trim=TRUE))
colnames (netflix_mov_year) <- "year"
#TV shows
netflix_tv0 <- read_html("https://www.finder.com/netflix-tv-shows/")
netflix_tv_year_0 <- html_nodes(netflix_tv0, 'td:nth-child(2)')
netflix_tv_year <- tibble (html_text (netflix_tv_year_0, trim=TRUE))
colnames (netflix_tv_year) <- "year"
Code for final count of the at Neflix ‘fresh’ and ‘old’ movies / TV shows portions is almost the same as for Disney
netflix_year <- rbind (netflix_mov_year, netflix_tv_year)
nrow (netflix_year)
netflix_year_table <- as.data.frame (sort (table (netflix_year), decreasing = TRUE), stringsAsFactors = FALSE) colnames (netflix_year_table) <- c('years', 'count')
glimpse (netflix_year_table)
Rows: 68
Columns: 2
$ years 2018, 2019, 2017, 2016, 2015, 2014, 2020, 2013, 2012, 2010,
$ count 971, 882, 821, 653, 423, 245, 198, 184, 163, 127, 121, 108,
Count 'fresh' and 'old' % in entire offer
netflix_year_table$years <- netflix_year_table$years %>% as.numeric () fresh_netflix_years <- netflix_year_table$years %in% c(2018:2020) fresh_netflix_num <- paste (round (sum (netflix_year_table$count[fresh_netflix_years]) / sum (netflix_year_table$count)*100),'%') old_netflix_years <- netflix_year_table$years < 2000 old_netflix_years <- na.omit (old_netflix_years) old_netflix_num <- paste (round (sum (netflix_year_table$count[old_netflix_years]) / sum (netflix_year_table$count)*100),'%')So, finally, I can build my arguments based on numbers, freshly baked and bold

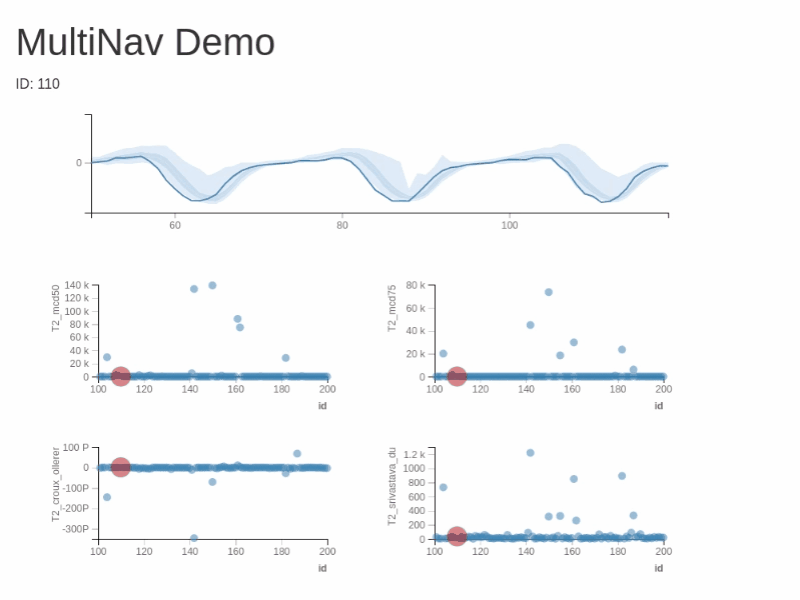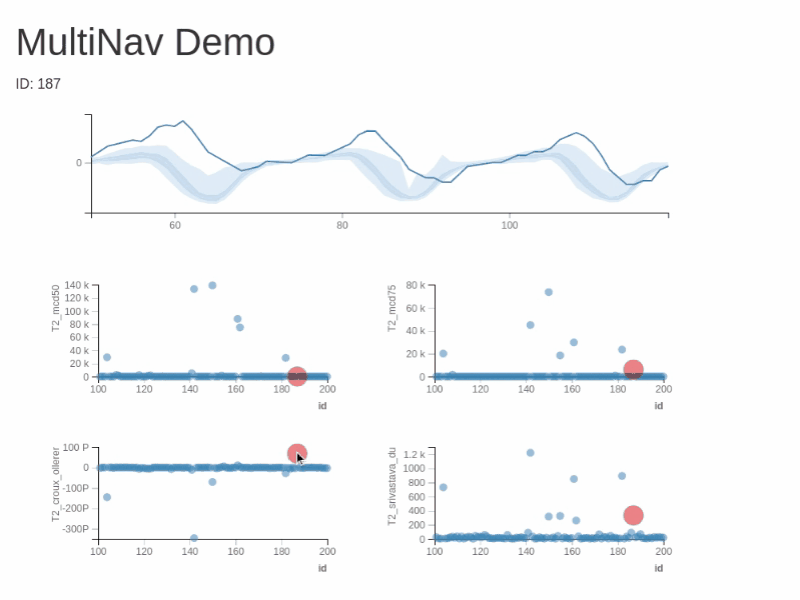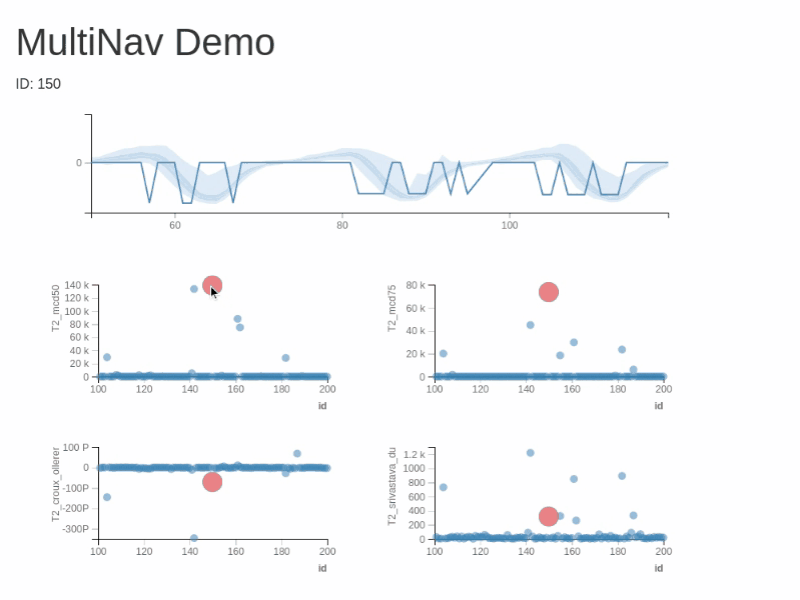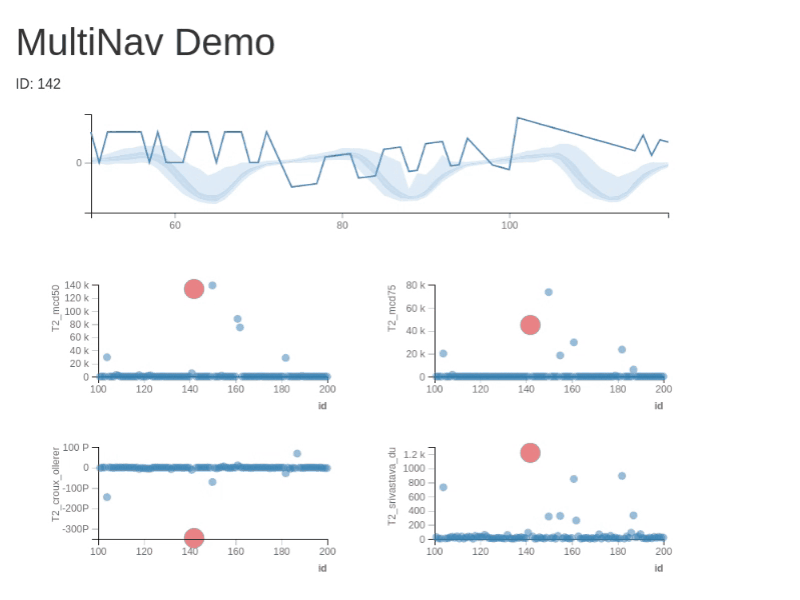



 You can test RXSpreadsheet by installing the GitHub version with devtools::install_github(‘MichaelHogers/RXSpreadsheet’) and subsequently running RXSpreadsheet::runExample().
This wrapper was made by Jiawei Xu and Michael Hogers. We are aiming for a CRAN release over the coming weeks. Special thanks to NPL Markets for encouraging us to release open source software.
You can test RXSpreadsheet by installing the GitHub version with devtools::install_github(‘MichaelHogers/RXSpreadsheet’) and subsequently running RXSpreadsheet::runExample().
This wrapper was made by Jiawei Xu and Michael Hogers. We are aiming for a CRAN release over the coming weeks. Special thanks to NPL Markets for encouraging us to release open source software. 
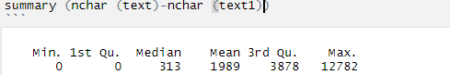
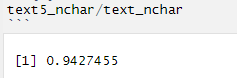
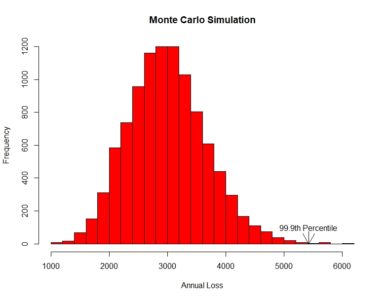 [contact-form][contact-field label=”Name” type=”name” required=”true” /][contact-field label=”Email” type=”email” required=”true” /][contact-field label=”Website” type=”url” /][contact-field label=”Message” type=”textarea” /][/contact-form]
[contact-form][contact-field label=”Name” type=”name” required=”true” /][contact-field label=”Email” type=”email” required=”true” /][contact-field label=”Website” type=”url” /][contact-field label=”Message” type=”textarea” /][/contact-form]