Learn how to use RMarkdown and Quarto! Join our workshop on RMarkdown and Quarto – Mastering the Basics which is a part of our workshops for Ukraine series.
Here’s some more info:
Title: RMarkdown and Quarto – Mastering the Basics
Date: Thursday, May 25th, 18:00 – 20:00 CEST (Rome, Berlin, Paris timezone)
Speaker: Indrek Seppo, a seasoned R programming expert, brings over 20 years of experience from the academic, private, and public sectors to the table. With more than a decade of teaching R under his belt, Indrek’s passionate teaching style has consistently led his courses to top the student feedback charts and has inspired hundreds of upcoming data analysts to embrace R (and Baby Shark).
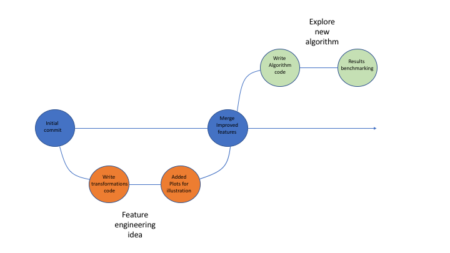
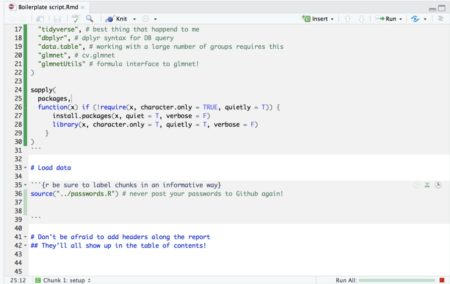
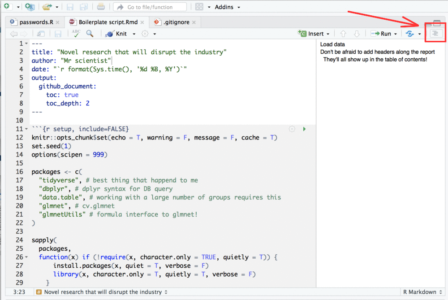
Description: Discover the power of RMarkdown and its next-generation counterpart, Quarto, to create stunning reports, slides, dashboards, and even entire books—all within the RStudio environment. This session will cover the fundamentals of markdown, guiding you through the process of formatting documents and incorporating R code, tables, and graphs seamlessly. If you’ve never explored these tools before, prepare to be amazed by their capabilities. Learn how to generate reproducible reports and research with ease, enhancing your productivity and efficiency in the world of data analysis.
Minimal registration fee: 20 euro (or 20 USD or 800 UAH)
How can I register?
- Go to https://bit.ly/3wvwMA6 or https://bit.ly/3PFxtNA and donate at least 20 euro. Feel free to donate more if you can, all proceeds go directly to support Ukraine.
- Save your donation receipt (after the donation is processed, there is an option to enter your email address on the website to which the donation receipt is sent)
- Fill in the registration form, attaching a screenshot of a donation receipt (please attach the screenshot of the donation receipt that was emailed to you rather than the page you see after donation).
If you are not personally interested in attending, you can also contribute by sponsoring a participation of a student, who will then be able to participate for free. If you choose to sponsor a student, all proceeds will also go directly to organisations working in Ukraine. You can either sponsor a particular student or you can leave it up to us so that we can allocate the sponsored place to students who have signed up for the waiting list.
How can I sponsor a student?
- Go to https://bit.ly/3wvwMA6 or https://bit.ly/3PFxtNA and donate at least 20 euro (or 17 GBP or 20 USD or 800 UAH). Feel free to donate more if you can, all proceeds go to support Ukraine!
- Save your donation receipt (after the donation is processed, there is an option to enter your email address on the website to which the donation receipt is sent)
- Fill in the sponsorship form, attaching the screenshot of the donation receipt (please attach the screenshot of the donation receipt that was emailed to you rather than the page you see after the donation). You can indicate whether you want to sponsor a particular student or we can allocate this spot ourselves to the students from the waiting list. You can also indicate whether you prefer us to prioritize students from developing countries when assigning place(s) that you sponsored.
If you are a university student and cannot afford the registration fee, you can also sign up for the waiting list here. (Note that you are not guaranteed to participate by signing up for the waiting list).
You can also find more information about this workshop series, a schedule of our future workshops as well as a list of our past workshops which you can get the recordings & materials here.
Looking forward to seeing you during the workshop!