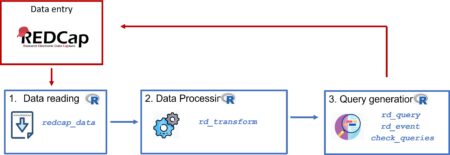
Data preprocessing is a critical step in any machine learning workflow. It ensures that your data is clean, consistent, and ready for modeling. In R, the recipes package provides a powerful and flexible framework for defining and applying preprocessing steps. In this blog post, we’ll explore how to use recipes to preprocess data for machine learning, step by step.
Here’s what we’ll cover in this blog: 1. Introduction to the `recipes` Package - What is the `recipes` package, and why is it useful? 2. Why Preprocess Data? - The importance of centering, scaling, and encoding in machine learning. 3. Step-by-Step Preprocessing with `recipes` - How to create a preprocessing recipe. - Centering and scaling numeric variables. - One-hot encoding categorical variables. 4. Applying the Recipe - How to prepare and apply the recipe to training and testing datasets. 5. Example: Preprocessing in Action - A practical example of preprocessing a dataset. 6. Why Use `recipes`? - The advantages of using the `recipes` package for preprocessing. 7. Conclusion - A summary of the key takeaways and next steps.
What is the recipes Package?
The recipes package is part of the tidymodels ecosystem in R. It allows you to define a series of preprocessing steps (like centering, scaling, and encoding) in a clean and reproducible way. These steps are encapsulated in a “recipe,” which can then be applied to your training and testing datasets.
Why Preprocess Data?
Before diving into the code, let’s briefly discuss why preprocessing is important:
-
Centering and Scaling:
-
Many machine learning algorithms (e.g., SVM, KNN, neural networks) are sensitive to the scale of features. If features have vastly different scales, the model might give undue importance to features with larger magnitudes.
-
Centering and scaling ensure that all features are on a comparable scale, improving model performance and convergence.
-
-
One-Hot Encoding:
-
Machine learning algorithms typically require numeric input. Categorical variables need to be converted into numeric form.
-
One-hot encoding converts each category into a binary vector, preventing the model from assuming an ordinal relationship between categories.
-
Step-by-Step Preprocessing with recipes
Let’s break down the following code to understand how to preprocess data using the recipespackage:
preprocess_recipe <- recipe(target_variable ~ ., data = training_data) %>% step_center(all_numeric(), -all_outcomes()) %>% step_scale(all_numeric(), -all_outcomes()) %>% step_dummy(all_nominal(), -all_outcomes(), one_hot = TRUE)
1. Creating the Recipe Object
preprocess_recipe <- recipe(target_variable ~ ., data = training_data)
-
Purpose: Creates a
recipeobject to define the preprocessing steps. -
target_variable ~ .: Specifies thattarget_variableis the target (dependent) variable, and all other variables intraining_dataare features (independent variables). -
data = training_data: Specifies the training dataset to be used.
2. Centering Numeric Variables
step_center(all_numeric(), -all_outcomes())
-
Purpose: Centers numeric variables by subtracting their mean, so that the mean of each variable becomes 0.
-
all_numeric(): Selects all numeric variables. -
-all_outcomes(): Excludes the target variable (target_variable), as it does not need to be centered.
3. Scaling Numeric Variables
step_scale(all_numeric(), -all_outcomes())
-
Purpose: Scales numeric variables by dividing them by their standard deviation, so that the standard deviation of each variable becomes 1.
-
all_numeric(): Selects all numeric variables. -
-all_outcomes(): Excludes the target variable (target_variable), as it does not need to be scaled.
4. One-Hot Encoding for Categorical Variables
step_dummy(all_nominal(), -all_outcomes(), one_hot = TRUE)
-
Purpose: Converts categorical variables into binary (0/1) variables using one-hot encoding.
-
all_nominal(): Selects all nominal (categorical) variables. -
-all_outcomes(): Excludes the target variable (target_variable), as it does not need to be encoded. -
one_hot = TRUE: Specifies that one-hot encoding should be used.
Applying the Recipe
Once the recipe is defined, you can apply it to your data:
# Prepare the recipe with the training data prepared_recipe <- prep(preprocess_recipe, training = training_data, verbose = TRUE) # Apply the recipe to the training data train_data_preprocessed <- juice(prepared_recipe) # Apply the recipe to the testing data test_data_preprocessed <- bake(prepared_recipe, new_data = testing_data)
-
prep(): Computes the necessary statistics (e.g., means, standard deviations) from the training data to apply the preprocessing steps. -
juice(): Applies the recipe to the training data. -
bake(): Applies the recipe to new data (e.g., the testing set).
Example: Preprocessing in Action
Suppose the training_data dataset looks like this:
| target_variable | feature_1 | feature_2 | category |
|---|---|---|---|
| 150 | 25 | 50000 | A |
| 160 | 30 | 60000 | B |
| 140 | 22 | 45000 | B |
Preprocessed Data
-
Centering and Scaling:
-
feature_1andfeature_2are centered and scaled.
-
-
One-Hot Encoding:
-
categoryis converted into binary variables:category_Aandcategory_B.
-
The preprocessed data might look like this:
| target_variable | feature_1_scaled | feature_2_scaled | category_A | category_B |
|---|---|---|---|---|
| 150 | -0.5 | 0.2 | 1 | 0 |
| 160 | 0.5 | 0.8 | 0 | 1 |
| 140 | -1.0 | -0.5 | 0 | 1 |
Why Use recipes?
The recipes package offers several advantages:
-
Reproducibility: Preprocessing steps are clearly defined and can be reused.
-
Consistency: The same preprocessing steps are applied to both training and testing datasets.
-
Flexibility: You can easily add or modify steps in the preprocessing pipeline.
Conclusion
Data preprocessing is a crucial step in preparing your data for machine learning. With the recipespackage in R, you can define and apply preprocessing steps in a clean, reproducible, and efficient way. By centering, scaling, and encoding your data, you ensure that your machine learning models perform at their best.
Ready to try it out? Install the recipes package and start preprocessing your data today!
install.packages("recipes")
library(recipes)
Happy coding! 😊