Interested in publishing a one-time post on R-bloggers.com? Press here to learn how.
It is usually said, that
for– and
while-loops should be avoided in R. I was curious about just how the different alternatives compare in terms of speed.
The first loop is perhaps the worst I can think of – the return vector is initialized without type and length so that the memory is constantly being allocated.
use_for_loop <- function(x){
y <- c()
for(i in x){
y <- c(y, x[i] * 100)
}
return(y)
}
The second
for loop is with preallocated size of the return vector.
use_for_loop_vector <- function(x){
y <- vector(mode = "double", length = length(x))
for(i in x){
y[i] <- x[i] * 100
}
return(y)
}
I have noticed I use
sapply() quite a lot, but I think not once have I used
vapply() We will nonetheless look at both
use_sapply <- function(x){
sapply(x, function(y){y * 100})
}
use_vapply <- function(x){
vapply(x, function(y){y * 100}, double(1L))
}
And because I am a
tidyverse-fanboy we also loop at
map_dbl().
library(purrr)
use_map_dbl <- function(x){
map_dbl(x, function(y){y * 100})
}
We test the functions using a vector of random doubles and evaluate the runtime with
microbenchmark.
x <- c(rnorm(100))
mb_res <- microbenchmark::microbenchmark(
`for_loop()` = use_for_loop(x),
`for_loop_vector()` = use_for_loop_vector(x),
`purrr::map_dbl()` = use_map_dbl(x),
`sapply()` = use_sapply(x),
`vapply()` = use_vapply(x),
times = 500
)
The results are listed in table and figure below.
| expr |
min |
lq |
mean |
median |
uq |
max |
neval |
| for_loop() |
8.440 |
9.7305 |
10.736446 |
10.2995 |
10.9840 |
26.976 |
500 |
| for_loop_vector() |
10.912 |
12.1355 |
13.468312 |
12.7620 |
13.8455 |
37.432 |
500 |
| purrr::map_dbl() |
22.558 |
24.3740 |
25.537080 |
25.0995 |
25.6850 |
71.550 |
500 |
| sapply() |
15.966 |
17.3490 |
18.483216 |
18.1820 |
18.8070 |
59.289 |
500 |
| vapply() |
6.793 |
8.1455 |
8.592576 |
8.5325 |
8.8300 |
26.653 |
500 |
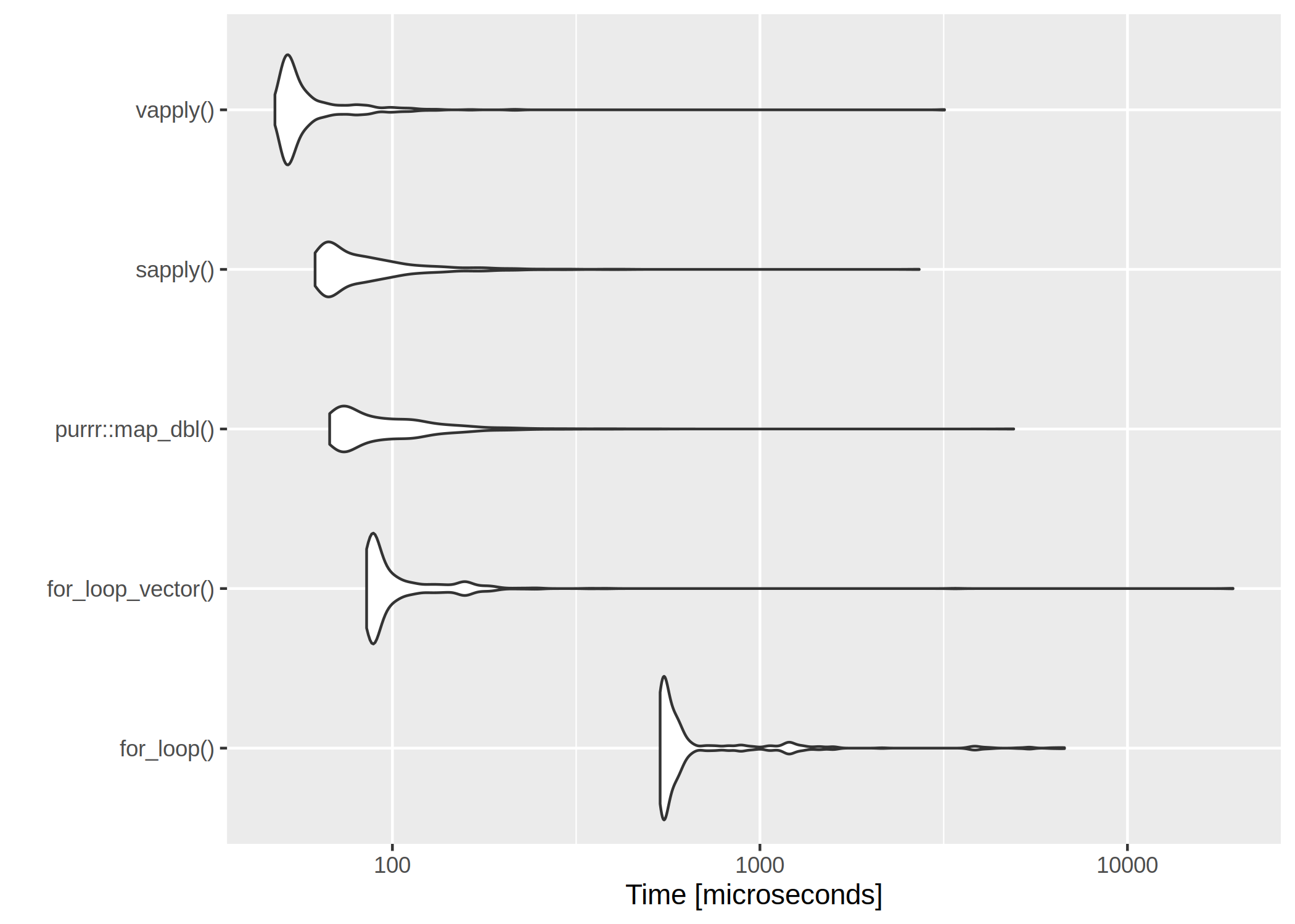
The clear winner is
vapply() and
for-loops are rather slow. However, if we have a very low number of iterations, even the worst
for-loop isn’t too bad:
x <- c(rnorm(10))
mb_res <- microbenchmark::microbenchmark(
`for_loop()` = use_for_loop(x),
`for_loop_vector()` = use_for_loop_vector(x),
`purrr::map_dbl()` = use_map_dbl(x),
`sapply()` = use_sapply(x),
`vapply()` = use_vapply(x),
times = 500
)
| expr |
min |
lq |
mean |
median |
uq |
max |
neval |
| for_loop() |
5.992 |
7.1185 |
9.670106 |
7.9015 |
9.3275 |
70.955 |
500 |
| for_loop_vector() |
5.743 |
7.0160 |
9.398098 |
7.9575 |
9.2470 |
40.899 |
500 |
| purrr::map_dbl() |
22.020 |
24.1540 |
30.565362 |
25.1865 |
27.5780 |
157.452 |
500 |
| sapply() |
15.456 |
17.4010 |
22.507534 |
18.3820 |
20.6400 |
203.635 |
500 |
| vapply() |
6.966 |
8.1610 |
10.127994 |
8.6125 |
9.7745 |
66.973 |
500 |
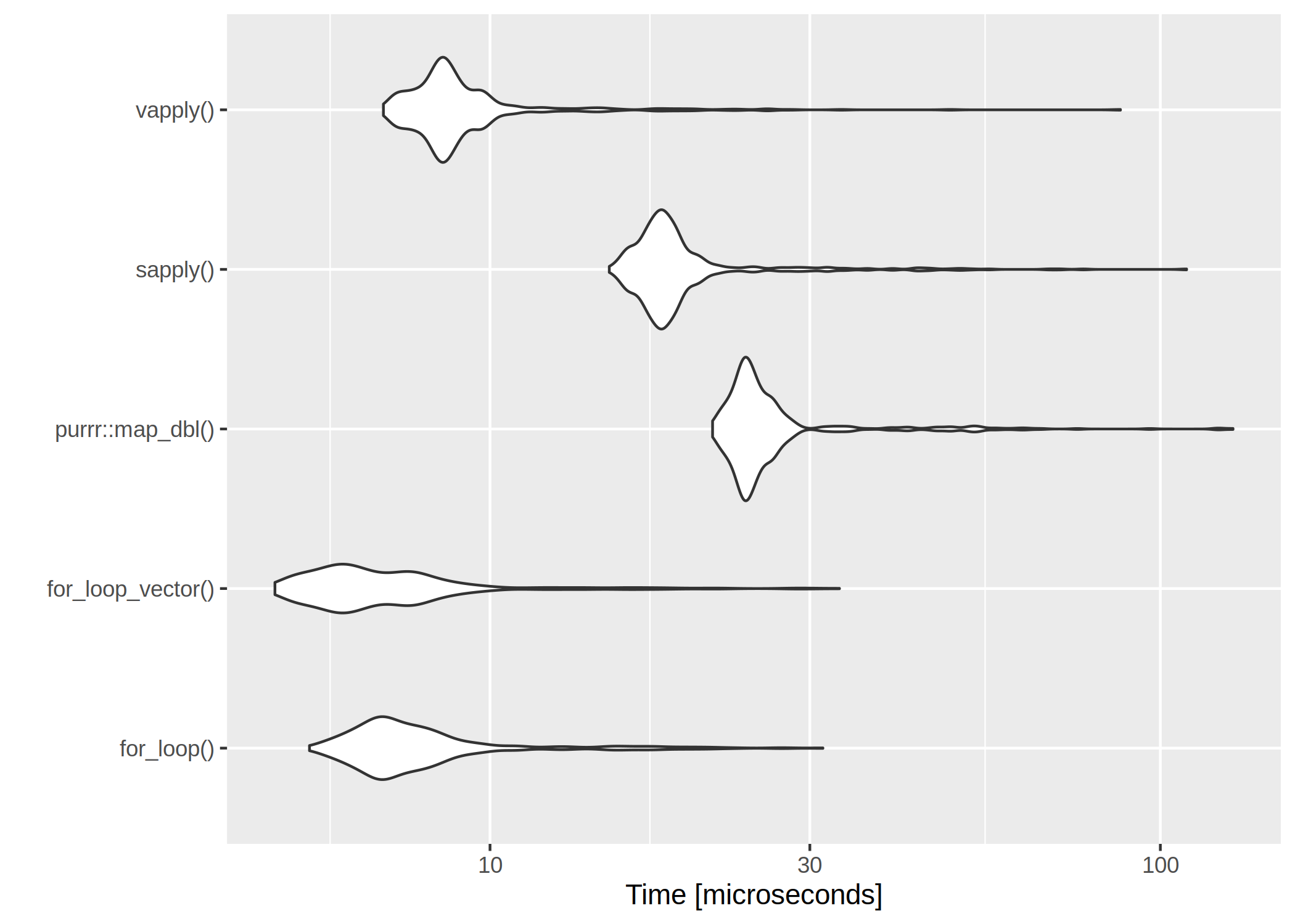
 The clear winner is vapply() and for-loops are rather slow. However, if we have a very low number of iterations, even the worst for-loop isn’t too bad:
The clear winner is vapply() and for-loops are rather slow. However, if we have a very low number of iterations, even the worst for-loop isn’t too bad:

Cool post! you might be interested into this Stack overflow question : https://stackoverflow.com/questions/48793273/why-not-use-a-for-loop/48793370#48793370 . Some argued that for loops could be faster but I couldn’t find any example of it happening. From your post it looks like for loops are faster when using a small amount of iterations. I’ll probably update my answer there with the insights gained here when I get the chance.
Thanks for the link. I think the mantra that for-loops should be avoided still holds. They might be slightly faster with few iterations, but the effect is not big, and will not affect the overall runtime.
Interesting (and possibly useful) results. However, your first table & graph seem to be inconsistent when it comes to the for loop results: As I try to look carefully at the numbers in the table and the resulting graph, it seems that, according to the table, for loops aren’t that bad; according to the graph, they are quite bad. I’m having trouble reconciling the two.
I suppose it is the difference between plotting all obtained runtime values and summary
I don’t think so. Your table shows a mean time for for_loop() of 11ms, with a max time of 71ms. Your figure looks like the minimum times are 700ms. It’s close to a two orders of magnitude difference.
to keep apples, apples you might want to re-run the sapply() one with USE.NAMES = FALSE (it shaves off some CPU cycles); also note that sapply() is rly just lapply() followed by a call to simplify2array() so another benchmark to possibly run is lapply() followed by unlist() with recursive = FALSE, use.names = FALSE.
Thanks. I was thinking of comparing map and lapply followed by flatten and unlist, but wanted to keep the comparison focused.
I’ll see if I can update the post
I think you don’t want to say for (i in x), but rather for (i in seq_along(x)).
You are right. Poor syntax
It is not a matter of simple “poor syntax”, your for loop return wrong results making wrong computation:
“`
identical(
use_for_loop(x),
use_vapply(x)
)
“`
Moreover they are of different length too!
“`
all.equal(
use_for_loop(x),
use_vapply(x)
)
“`
using `for (i in x)` you are running along the element of x and not along their position, i.e. let `x <- 1000`, when you call
“`
y[1] <- x[1] * 100
“`
you ara asking R to perform
“`
y[1000] <- x[1000] *100
“`
so, because of the single brackets are used to subsect and create vectors (and not to access to an element of them) R create for you the position 1000 in the vector y (if it has not already at least 1000 elements), next R create a subvector with the element in position 1000 of x, and if it not exists the result of this subsetting is NA.
so finally, your result for the for loop return a vactor with a random length in which at random position are NA, and on the other are random numbers. And the computation go through the creation and replacement of new vectors of random length at every step. It is not surprising it is slow… but the most important fact is that the results returned by the loop is wrong. And you do not recieve any error or warning!! (and here is one of the real value of using functions in the tidyverse…)
On the other hand, it would not been a simple metter of syntax (if you mean "style") anyway. This, because syntax matter in the computation.
“`
use_for_loop_good_vector <- function(x){
y <- vector(mode = "double", length = length(x))
for(i in seq_along(x)){
y[[i]] <- x[[i]] * 100
}
return(y)
}
“`
It is an order of magnitude quicker than you top winner iterator!!
and that is only thanks to the fact you loop through a sequence spanning on the indeces of x (and not trough the element of x, but we have already discussed this problem). And, to the fact that you want replace **the** element **in** the position i of y and you want to use **the** element **at** the position i of x so you have to use double square bracket here. With a single square bracket you ask to R to create a new vector mady by the element(s) of the original one indicated inside the bracket. With the double bracket, you access to the content of a single (!!) element of a vector, i.e. its value. So, the mantra should be translated in "poor for loops should be avoided in R" if you look at the performance.
Anyway, I would like to make a consideration: my version is ten time faster then the best one of yours…wooow, …. but I have saved 9 MICRO seconds!! How much time I would heve saved by typing `map(x, ~.*100)` instead of seven line of code? How much time I will lose whenever I will look back at the code to change a parte of it or to understand what I have coded?
More important, all your "vectorized" (implicit for loop) versions are not wrong (even if sapply can fail without warning nor errors for other reasons,
so I suggest to avoid it!).
That said, I agree with the original mantra "foor loop should be avoid in R", but for different reason:
– the execution time is not related to R but on you, mainly
– the time you spend to code it is much higher than the alternatives
– what you expect from the computation and how do you want to obtain it is clearer using the alternatives
– the possibility to obtain wrong results without any wornings or errors are much higher then the alternatives (and depends on tiny syntax/style/code details often difficult to debug… expecially if you have 7 full line of code doing that stuff)
So, please, do not "correct" your code. But, update it highlighting your tought about this for who after you will reed your post in the future.
You need to be careful what you’re measuring here. After the first call to a function, R’s JIT compiler kicks in and you’ll be running compiled versions of all of the functions, in addition to adding the overhead of a function call.
If you’re only interested in comparing how loops and apply functions compare, rather than how they compare after being byte-code compiled, you should turn off the compiler with
enableJIT(0)
before running your benchmarks.
Even better would be to eliminate the functions altogether and just wrap each item you want to compare in a set of curly braces. This also removed the overhead of the function calls from each.
Thank you for the hint, I was not aware of this.
I will see if I can improve and update the post.
As others hinted – the contents of your loop are so tiny that it’s likely that various startup and closeout tasks heavily bias all your results. I would recommend writing at least 10 or so lines of messy functions inside your loop. I would also recommend making the loop at least 10^4 cycles if not more.
It s not possible to modify the original post, so the improved and extended the run-time comparison is available as a gitlab snippet: https://gitlab.com/snippets/1869401
Hi Ulrik, I think you can update the post, and I’ll make sure to re-publish it manually to r-bloggers.
Sounds great!